Pathologists may soon have access to a new predictive tool for urinary tract infections, following the development of a machine-learning model that can forecast phenotypic antibiotic susceptibility with high accuracy using polymerase chain reaction (PCR) results and demographic information. The model, created by Pathnostics, provides patient-specific probabilities for which antibiotics are likely to be effective, potentially reducing empiric therapy and treatment failures. Jim Havrilla, PhD, senior director of data and AI at Pathnostics, presented the data Thursday at the Association for Molecular Pathology (AMP) annual meeting in Boston.
“The point of the Guidance UTI predictive report is to both reduce empiric therapy and treatment failure rates,” Dr. Havrilla said, describing the test that underpins the model. He emphasized that the predictive component provides actionable information in PCR turnaround time (<8h) while clinicians await full phenotypic susceptibility results, which are delivered within 24 to 36 hours.
Guidance UTI, Pathnostics’ flagship test, combines multiplex PCR for pathogen and resistance-gene detection with pooled antibiotic susceptibility testing (P-AST). The predictive model uses results from the PCR assay along with patient demographics—age, gender, and location—to generate probabilities for antibiotic effectiveness. “All the input data is from PCR and demographic data, and it’s trained on a truth set of our phenotypic data results,” Dr. Havrilla said.
The model currently encompasses 20 separate algorithms, one for each antibiotic on the panel, which are aggregated into a unified output. Only antibiotics with a predicted probability above 60% are reported, ensuring that clinicians receive guidance only when it is likely to be informative. “If it’s not better than a coin flip, we’re not going to even share it with you,” Dr. Havrilla explained.
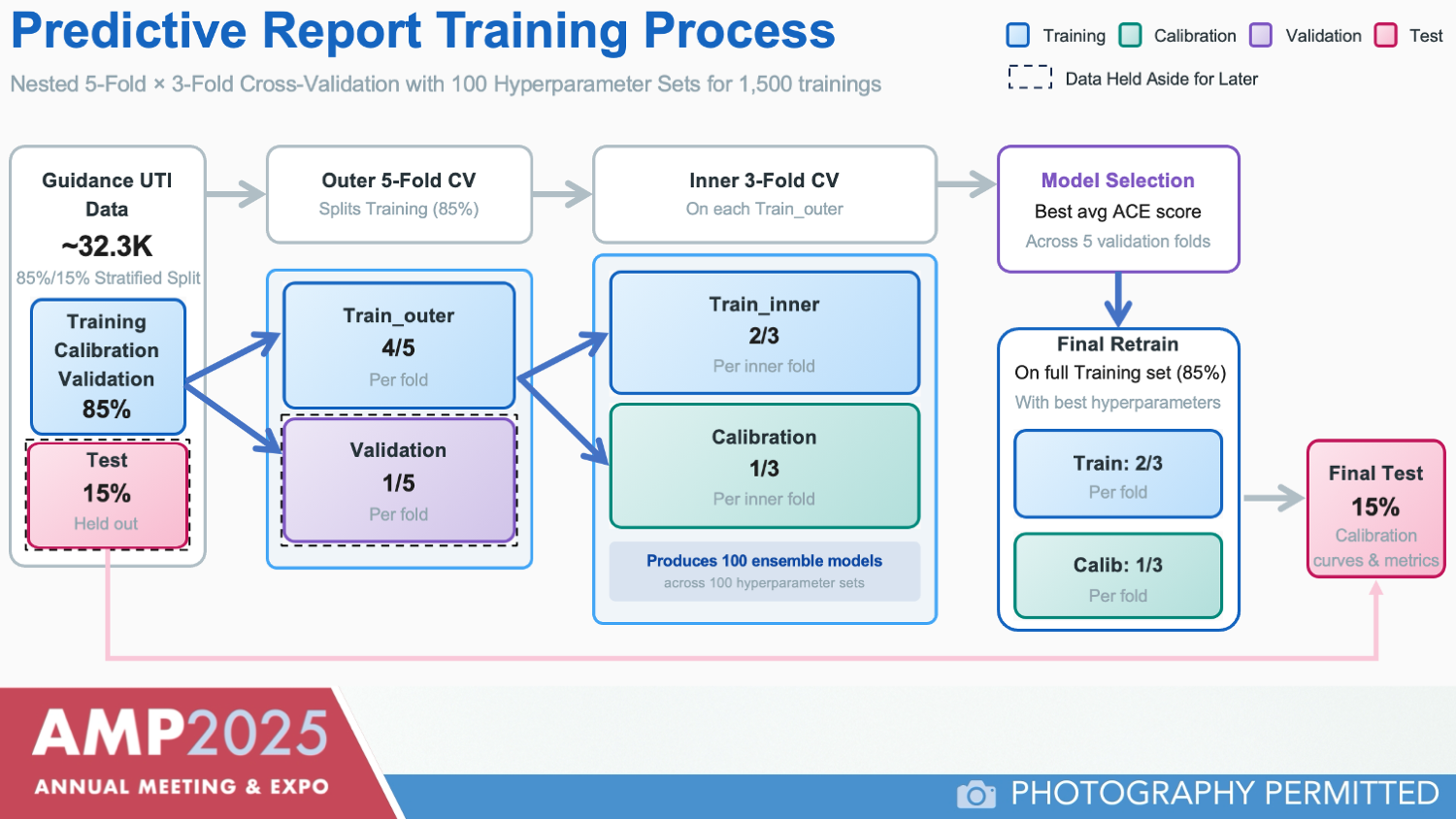
Dr. Havrilla outlined the model’s development using roughly 32,300 samples with paired phenotypic and PCR data (Figure 1). Data were split into training, calibration, validation, and test sets, and the model was developed using a light gradient boosting machine, a decision tree–based algorithm. Multiple hyperparameter configurations and nested cross-validation were employed to optimize calibration.
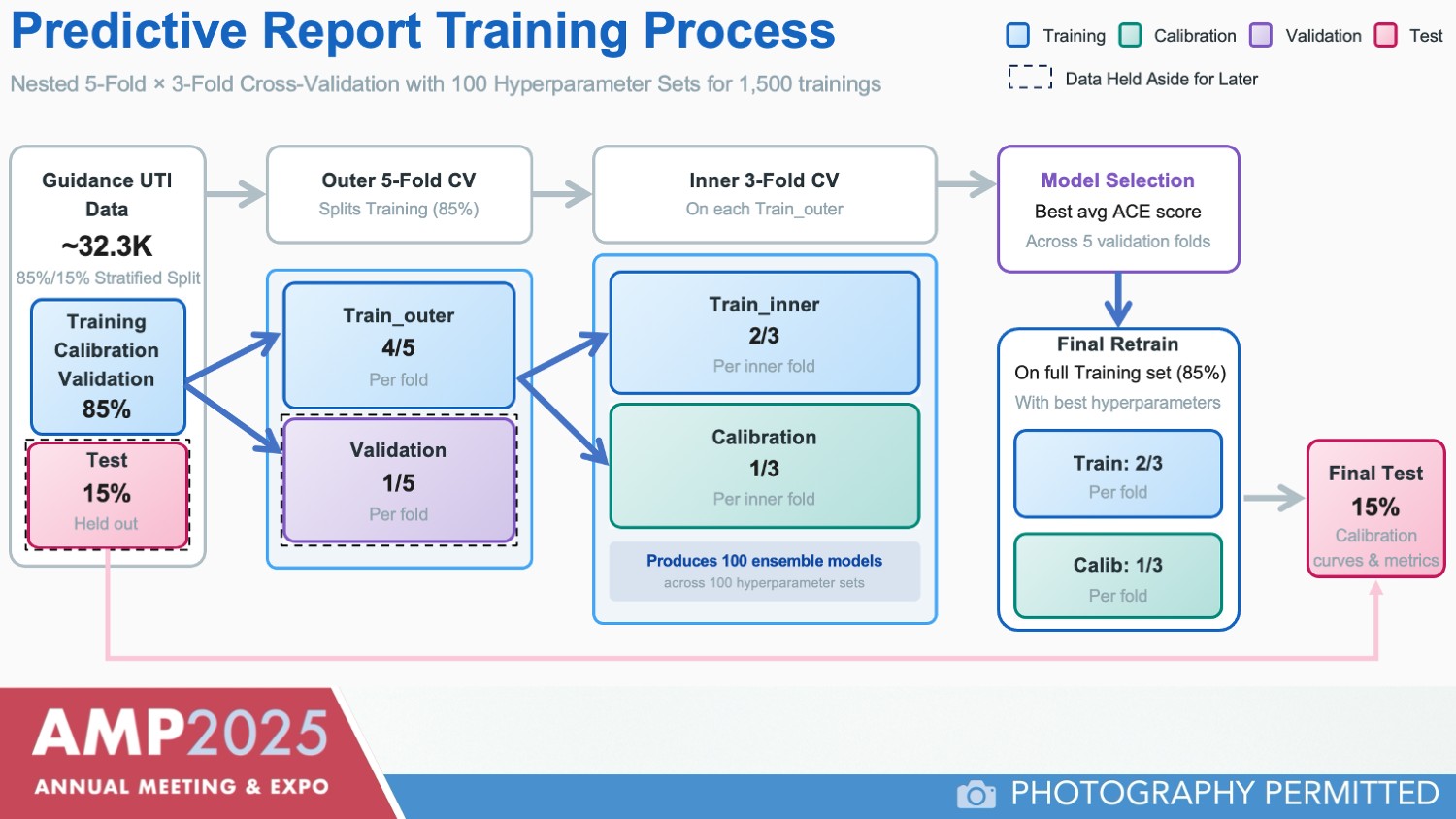
Calibration of the model is an important step toward ensuring predictive accuracy, emphasized Dr. Havrilla. “You want a curve where the probability that you see from your model’s output very closely matches the fraction of positives you see in your actual data,” he explained (Figure 2).
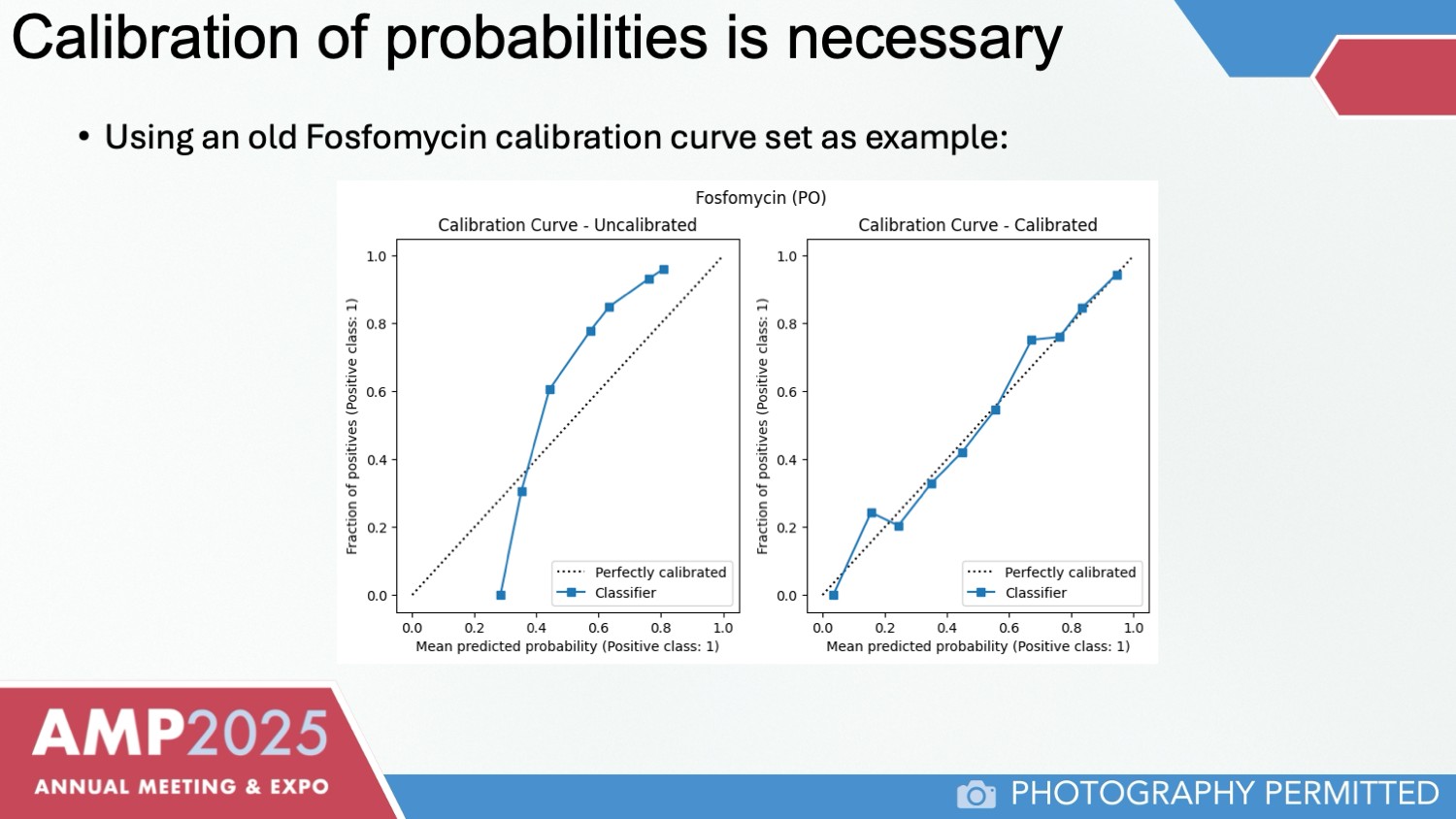
Initial results show that the model’s predictions align closely with phenotypic results, particularly for first-line antibiotics such as nitrofurantoin and fosfomycin, and high-sensitivity antibiotics like piperacillin-tazobactam, ampicillin-sulbactam, and amoxicillin-clavulanate. Confidence intervals are expected to narrow as additional data are incorporated (Figure 3).
Dr. Havrilla noted that the model continues to evolve: “We’re always iterating in the development space on newer versions of the model as data comes in.”
Pathnostics plans to roll out a pilot launch of the predictive model in 2026. Dr. Havrilla said the goal is to provide rapid, actionable insights to clinicians managing complex or recurrent UTIs, improving patient outcomes and optimizing antibiotic use.