Researchers at the University of Chicago have developed a method to extract and analyze genetic information from cancer samples that are nearly a century old, an advance that could reshape how scientists understand the evolution of disease. By adapting techniques used in archaeologic DNA research, the team successfully recovered genomic and microbial data from colorectal cancer tissue collected as far back as 1932.
The findings, presented Thursday by Alexander Guzzetta, MD, PhD, at the 2025 Association for Molecular Pathology (AMP) Annual Meeting in Boston, demonstrate that even long-degraded formalin-fixed, paraffin-embedded (FFPE) specimens can yield meaningful DNA for sequencing. The study, conducted with ancient DNA expert Maanasa Raghavan, PhD, shows that these archival materials—long thought unsuitable for genomic analysis—may offer a new window into how diseases change over time.
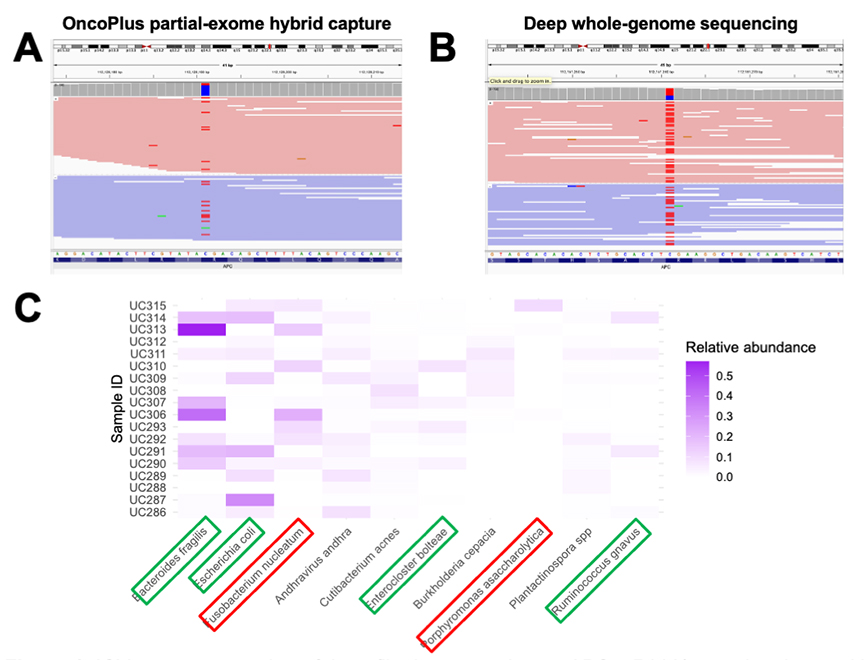
Dr. Guzzetta explained that his University of Chicago team analyzed colorectal cancer samples spanning nearly a century using optimized extraction and sequencing techniques (Figure 1). The researchers modified conventional genomic workflows to retain and interpret the smallest fragments of damaged DNA, allowing them to identify canonical driver mutations such as TP53 and APC even in samples collected before World War II. They also detected microbial DNA consistent with both normal gut flora and bacteria previously linked to colorectal cancer, including Fusobacterium nucleatum. The study’s methods could be used to help explain why colorectal cancer rates have risen sharply among younger adults, a trend that has puzzled clinicians for years.
“This approach could unlock the ability to study how diseases evolve over decades and shed light on how their biology has changed through time,” Dr. Guzzetta said.
Researchers hope the technique can be expanded to other diseases where historical samples exist but genomic data remain inaccessible. If widely adopted, the approach could transform archived pathology collections worldwide into vast datasets for understanding how human diseases have shifted alongside environmental, dietary, and microbial changes.